Machine Learning
Lanthanide-based Single Molecule Magnets (SMMs) hold promise for their remarkable magnetic properties, driven by significant inherent anisotropies. Understanding how the surrounding ligand structure affects their magnetic behavior involves working with a set of 27 parameters derived from ligand field theory. However, experimental data which is typically related to magnetization and susceptibility curves, is relatively featureless for these materials. The over-parameterized problem of determining model parameters from experimental data becomes challenging, as multiple parameter sets can characterize the data with equal validity.
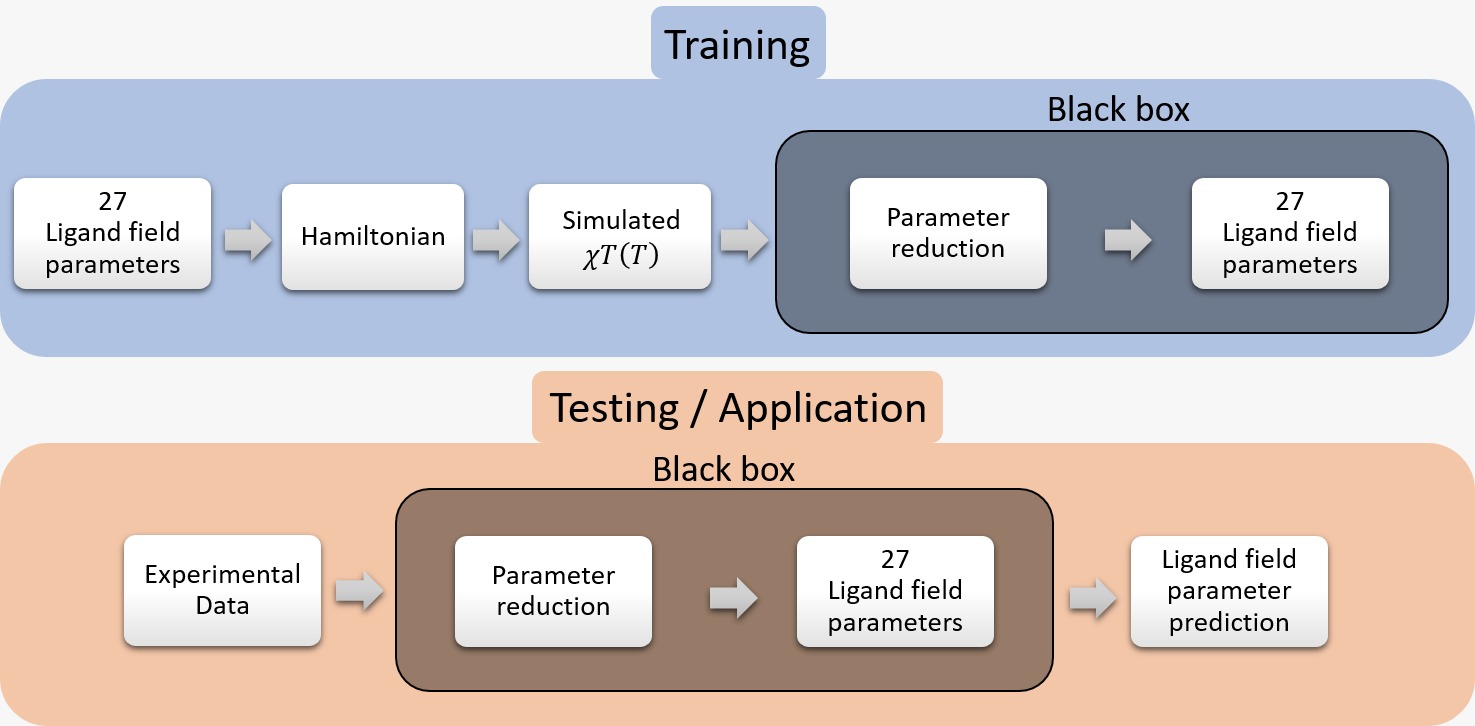
A possible machine learning approach tackles this problem by finding a new parameterization of the data which is unique and not physically/chemically motivated. This reduced parameterization is then related to the 27 ligand field parameters, which involves learning a one-to-many mapping. The two steps of parameter reduction and mapping constitute a “black-box” algorithm which, once trained, can take in measured experimental data and predict its corresponding sets of ligand field parameters.
The ill-posed mapping from the reduced parameters to the ligand field parameters is a general inverse problem which opens itself to an increasing variety of solution approaches from many different fields of research. In deep learning particularly, Variational Autoencoders (VAEs) and Invertible Neural Networks (INNs) have recently shown to be successful at modelling inverse functions.
Machine learning applied to molecular magnetism requires due consideration for the underlying physics, which can motivate the direction of research. However, the relatively uncharted nature of this work allows open-ended and fresh avenues of questioning, as well as formulation of the underlying problem in alternate ways that would motivate new approaches and techniques.